New research points to the shingles vaccine meaningfully reducing the number of people who develop either dementia or mild cognitive decline later in life https://econ.st/4s73gdz.
Illustration: Cristina Spanò
The evidence is surprisingly strong.
New research points to the shingles vaccine meaningfully reducing the number of people who develop either dementia or mild cognitive decline later in life https://econ.st/4s73gdz.
Illustration: Cristina Spanò
The evidence is surprisingly strong.

Among its numerous health benefits, physical activity reduces the risk of developing Alzheimer’s disease. A new study on mice now dives into the specific mechanisms and proteins that allow exercise to protect our brains.
Scientists had previously determined that physical activity increases a protein called glycosylphosphatidylinositol-specific phospholipase D1 in the blood of mice, and that this protein is associated with good brain health.
That protein – more succinctly referred to as GPLD1 – strengthens the barrier that guards the brain against all sorts of unwelcome visitors within our blood, protecting against inflammation and subsequent cognitive decline.
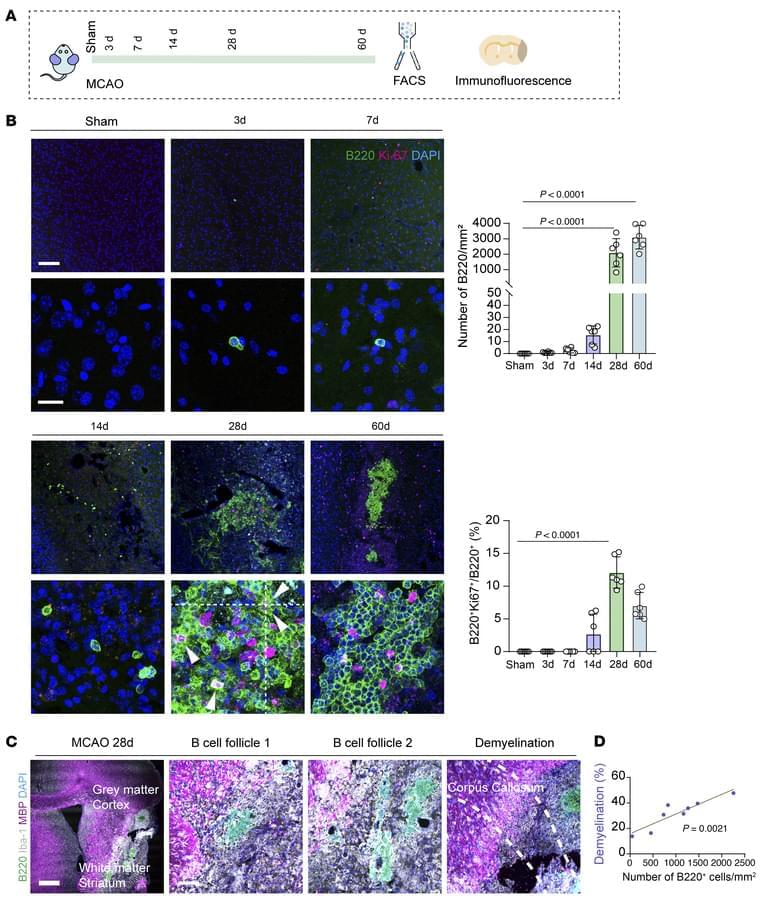
Here, Chuan Qin & team use complementary models in experimental ischemic stroke, showing early post-stroke stages in which microglia recruit B cells into ischemic lesions through MIF/CD74/CXCR4, while later stage post-stroke effects involve interferon signaling in B cells that drives neuroinflammation and brain injury:
The image shows B lymphocytes (Green) in mouse dura tissue colocalizing with CD31+ blood vessels (Red).
1Department of Neurology, Tongji Hospital, Tongji Medical College and State Key Laboratory for Diagnosis and Treatment of Severe Zoonotic Infectious Diseases;
2Key Laboratory of Vascular Aging, Ministry of Education, Tongji Hospital of Tongji Medical College; and.
3Hubei Key Laboratory of Neural Injury and Functional Reconstruction, Huazhong University of Science and Technology, Wuhan, Hubei, China.

Neocortical expansion driven by basal progenitors.
The emergence of indirect neurogenesis, driven by highly proliferative basal progenitors, contributed to the significant enlargement of the mammalian neocortex during brain evolution.
In recent years, several human-specific genes and enhancers have been described that differentially affect the biology of progenitor cells and potentially contribute to the increased neocortical complexity and disease-susceptibility of the human brain.
Emerging research is uncovering multiple pathways that disrupt basal progenitor biology, emphasizing these pathways’ involvement not only in classical neurogenesis-related disorders such as microcephaly but also in neurodevelopmental conditions traditionally linked to impairments in neuronal connectivity. sciencenewshighlights ScienceMission https://sciencemission.com/Basal-progenitors
The diversification and expansion of distinct progenitor cell subtypes during embryogenesis are essential to form the sophisticated brain structures present in vertebrates. In particular, the emergence of highly proliferative basal progenitors contributed to the evolutionary enlargement of the mammalian neocortex. Basal progenitors are at the center of indirect neurogenesis and can be divided into two main subtypes: the classical TBR2-positive intermediate progenitor cells and the outer radial glial cells, which are especially abundant in gyrencephalic species. While the function of some transcriptomic regulators is conserved across the mammalian clade, recent studies have identified human-specific genes and enhancers that uniquely affect progenitor biology, possibly driving the increased neocortical complexity and disease-susceptibility of the human brain.




Scientists have uncovered a surprising new role for little-known brain cells called tanycytes that may influence the development of Alzheimer’s disease. These specialized cells appear to help remove toxic tau protein from the brain by transporting it from the cerebrospinal fluid into the bloodstream. When tanycytes become damaged or dysfunctional, tau can accumulate in the brain, a hallmark of Alzheimer’s.

For centuries we treated technology as a tool, and now a new movement insists it is becoming the future of the human species itself.
Transhumanists like Harari and Kurzweil predict the merger of humans and machines, even the rise of a “digital God.” But critics fear this proposed future, calling transhumanism “the world’s most dangerous idea.”
Is the future one where technology is not merely a source of innovation but the basis for a new account of what it is to be human, or are claims of eternal life and new forms of intelligence just fanciful nonsense?
Joining the debate are transhumanist pioneer Zoltan Istvan, physicist and consciousness researcher Àlex Gómez-Marín, philosopher of mind Susan Schneider, and Softmax co-founder Adam Goldstein.
Tap the link now to watch the full debate.
We have for centuries sought technological progress. But now some are making the radical claim that technology is the future of the human race. ‘Effective accelerationists’ have won high-profile Silicon Valley support and claim we should accelerate technology to.
