Whereas cellular senescence is known to promote aging, many of the mechanisms controlling this process remain poorly understood. Using human mesenchymal precursor cells (hMPCs) carrying pathogenic mutations of the premature aging diseases Werner syndrome and Hutchinson-Gilford progeria syndrome, the authors conducted a genome-wide CRISPR-Cas9–based screen to identify genes that could affect cellular senescence. They identified KAT7, a histone acetyltransferase gene, as a driver of senescence. Inactivation of Kat7 in mice aging normally and in prematurely aging progeroid mice extended their life span. Although KAT7 requires further study in other cell types, these experiments highlight the utility of genome-wide CRISPR-Cas9 screens and shed further light on mechanisms controlling senescence.
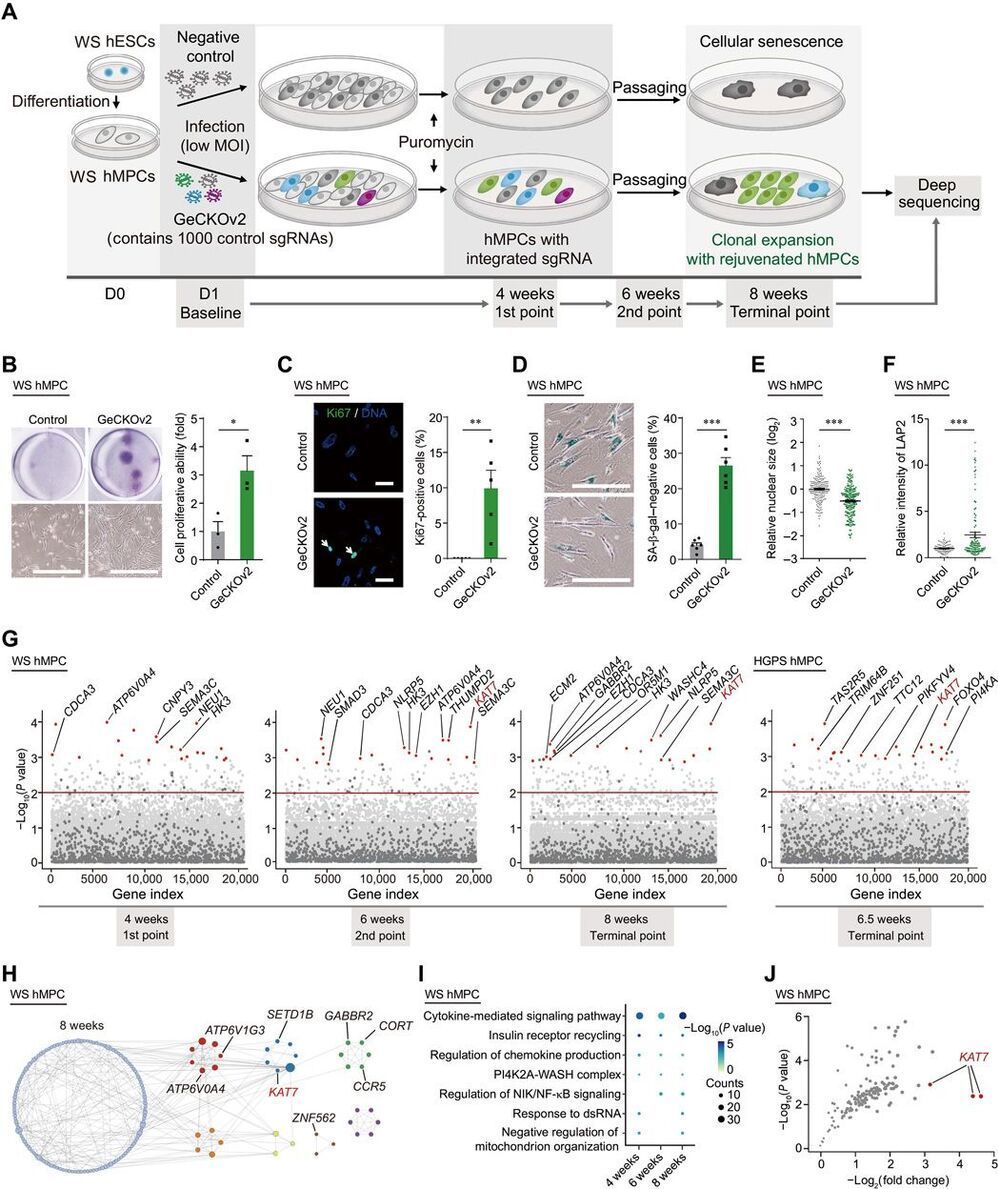
Understanding the genetic and epigenetic bases of cellular senescence is instrumental in developing interventions to slow aging. We performed genome-wide CRISPR-Cas9–based screens using two types of human mesenchymal precursor cells (hMPCs) exhibiting accelerated senescence. The hMPCs were derived from human embryonic stem cells carrying the pathogenic mutations that cause the accelerated aging diseases Werner syndrome and Hutchinson-Gilford progeria syndrome. Genes whose deficiency alleviated cellular senescence were identified, including KAT7, a histone acetyltransferase, which ranked as a top hit in both progeroid hMPC models. Inactivation of KAT7 decreased histone H3 lysine 14 acetylation, repressed p15INK4b transcription, and alleviated hMPC senescence.








Comments are closed.